GSoC 2024 - 2D visualization for small molecules
02 Aug 2024Contributor: Valerij Talagayev (@talagayev)
Mentors: Cédric Bouysset (@cbouy), Richard Gowers (@richardjgowers) and Yuxuan Zhuang (@yuxuanzhuang)
Organization: MDAnalysis
Release: GSoC 2024: 2D visualization for small molecules Release
During the Google Summer of Code 2024 program I was working on the 2D Visualization of small molecules, which is an important project, that would allow people to easily visualize the molecule that they have in their file
through the selection of the molecule as an AtomGroup in MDAnalysis.
Goals of the project
The goal of the project was to create a code, that would allow the user to select an AtomGroup with the help of MDAnalysis, which would be used with an RDKit Converter to convert the molecule into an rdkit mol object, which would be used for the visualization.
My Proposal consisted of the following steps:
- Select the AtomGroup via an input from the user via
u = mda.Universe("input.pdb")to create an MDAnalysis Universe andag = u.select_atoms("resname UNK")to select theAtomGroupwith the molecule that needs to be displayed - Convert the selected AtomGroup into an rdkit mol object via
ag.convert_to("RDKit") - Apply
Chem.RemoveHsandAllChem.Compute2DCoordsto obtain a 2D visualization of the molecule - Display the molecule via
IpyWidgetswith different checkboxes, that by selection would highlight specific parts of the molecule
After discussion with the mentors and suggestions regarding how the code should be structured, it was decided that I would proceed with this suggestion and would apply the following steps with later on adding the additional
features in the IpyWidgets visualization, which would give the user important information in regards to the characteristics of the molecule that is being visualized.
What I did
The outcome of this GSoC program is a package called mdonatello which was created as follow:
Main Visualization Code
The main part of the code consisted of setting up the widget for the visualization, which would allow the display of the molecule in a jupyter notebook. This is performed in the
MoleculeVisualizer class that is responsible for the visualization.
It initializes several interactive widgets from IpyWidgets such as a dropdown menu to select the molecule that the user wants to display and the
checkboxes that control the aspect of the image displayed, as explained in the next section.
This main code was the first part of the code that I designed and was in the beginning only able to display the molecule and had only one interactive checkbox, which displayed the atom indices of the molecules, but as more and more features were added the visualization grew bigger and had more features that it needed to display.
Feature Addition
The feature addition was the next step that I was working on and was mainly the biggest part of the project since there was always an additional interesting feature that could be added that was proposed either by myself or one of the mentors.
The first feature addition that I was working on were the physiochemical properties as well as the number of hydrogen bond donors and acceptors of the molecule.
Next, I added checkboxes that would highlight the related features on the image. For example, I implemented pharmacophore checkboxes,
which by selection would highlight the selected pharmacophores, thus by selecting a hydrophobic feature, the atoms of the molecule that are hydrophobic via the RDKit pharmacophore
recognition were highlighted in the assigned colors.
Here is a short summary of features that I added for the visualization, which would show a value for the corresponding feature or adjust the figure of the molecule to display the values:
- Phyisochemical Properties
- Atom Indices
- Partial Charges
- Hydrogen Bond Donors&Acceptors
And here is a short summary of features, that will highlight a specific atoms and bonds corresponding to their features if they are selected:
- Rotatable Bonds
- Partial Charge Heatmap
- Functional Groups
- Stereocenters
- Murcko Scaffold
- Pharmacophores
There is a separate checkbox for each pharmacophoric feature, thus you can select the specific feature that needs to be highlighted.
For the functional groups it currently uses SMART Patterns to identify certain functional groups and will then generate the checkboxes of the functional groups that were recognized and by selecting them you are able to see the atoms of this functional group.
Code Restructure
This is also a section where the work that was done throughout the whole project. In the beginning, the code was mainly consisting of functions in one file and this was one of the points that I was glad that my mentors helped me with. The code restructure allowed me to go from a code that was first based upon only function to structure it with classes. In the end, the separate classes were moved into separate files. The benefit of such a structure is that by having each class/function have a single responsibility, the code is divided into small parts, which makes it easier to understand the code and maintain it.
Actions, Testing and Documentation
This would be the final part of what I did and here I would put all these three parts together. A testsuite based on the pytest framework was created for this project, in addition to the code being uploaded to
Codecov to display the coverage of the code. Furthermore, a CI CD workflow was created, which installs the package and runs the tests on
Linux, MacOS and Windows systems with the Python versions 3.10, 3.11 and 3.12 with different IpyWidgets versions, with the oldest supported version for this code being 7.6.4. Finally,
a documentation based on the MDAKits Template was created and can be found here: https://mdonatello.readthedocs.io/
Current State
MDonatello is available and can be installed and used on your system (see the How to use it section). Some improvements are planned as explained below.
What’s left to do
The code is working, but there is always things that can be improved. The main parts of the code that would need to be improved would be the functional groups, since currently the SMART
Patterns are not optimal and it leads to cases of functional groups overlapping, an example being the recognition of an hydroxyl group in an carboxylic acid group, thus such cases need to
be separated. Additionally, the figure generation is not optimal, since currently it uses Draw.MolToImage, which during the discussion with the mentors was decided as an not so good option
and it would need to be adjusted. Additionally, the Partial Charge Heatmap is currently mainly highlighting the atoms, but there is an option to display it as a heatmap, which would require
code adjustment. Then the update_display is also not optimal currently and would need to be improved. There are also some small code structure details, that would need to be adjusted for
the code to be more clean and structured and last, but not least the code would need to be moved into the MDAKits and also published in conda-forge and pypi to make it more accessible.
What code got merged (or not) upstream
There were multiple PRs created for this project, but they were merged to obtain a big and summarized PR, which is also the version that is used in the Release
How to use it
Currently MDonatello is a separate package that can be git cloned and installed with the following steps:
First clone the repository:
git clone https://github.com/talagayev/MDonatello
Create a virtual environment and activate it:
conda create --name mdonatello
conda activate mdonatello
Then go into the MDonatello folder:
cd MDonatello
Finally this package from source:
pip install -e .
To use the mdonatello package you need to run a jupyter notebook, thus run the command:
jupyter notebook
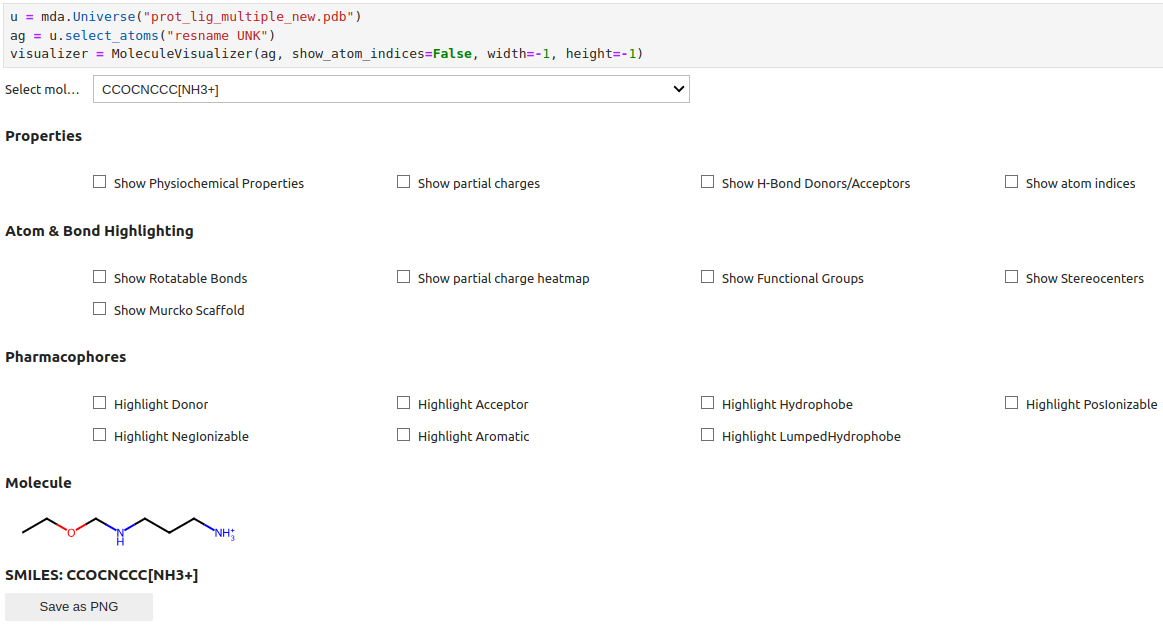
Now that you started a jupyter notebook create a notebook file and enter the following command to use mdonatello: Here you need to adjust the name of the PDB File to your PDB File and the resname of the molecule to your molecule
import MDAnalysis as mda
from mdonatello import MoleculeVisualizer
u = mda.Universe("input.pdb")
ag = u.select_atoms("resname UNK")
visualizer = MoleculeVisualizer(ag, show_atom_indices=False, width=-1, height=-1)
This would lead you to obtain the following display, where you could then select the molecule and checkboxes:
Lessons Learned During GSoC
Here I would highlight some lessons that I learned during GSoC2024:
- Have an Idea of the overall structure of the code
- Discuss the ideas with the mentors to see their opinion
- Don’t be afraid to ask questions
- Try to work on different aspects of the code (in my case I was almost always only focusing on feature addition and needed to get reminded that there are other parts)
- Try to learn as much from the mentors and apply it in your code
- Getting a meeting with all mentors that have different time zones is sometimes tricky :)
Conclusion & The Future
It was a fun project and I liked working on it, but the work doesn’t stop here. As I mentioned some aspects of the code can be improved and things that can be done, so I will continue to work on the code after GSoC 2024, here are again the short highlights that I already mentioned that I would work on:
- Code structure improvement
- Figure Generation and SMART Patterns need to be improved
- Heatmap needs to be created for partial charges instead of atom highlighting
- Adding the package to
MDAKitsandconda-forge - New Features can be added
Acknowledgements
I would like to thank MDAnalysis for giving me the opportunity to work on this project. The application process was very nice and detailed, I liked it that during the application process. I was able to get to know the MDAnalysis code more and contribute to the code and get to know some people from the community during this time.
I would like to thank Oliver Beckstein (@orbeckst) and Jenna M Swarthout Goddard(@jennaswa) for the insights and help with the organization and structure during the application, I am glad that I was able to contribute to MDAnalysis with this project.
I would also like to thank my mentors Cédric Bouysset (@cbouy), Richard Gowers (@richardjgowers) and Yuxuan Zhuang (@yuxuanzhuang), who helped me a lot during the project, with their helpful and insightful mentoring. I am glad, that I was able to learn from you and it helped me to improve my coding skills. Also, shoutout to Hugo MacDermott-Opeskin (@hmacdope) for his help with the PR requests during the application process.
Finally, I would also like to thank Google for offering this program and supporting open-source software.