Contributing new datasets
New datasets are very welcome and everybody is encouraged to make their
datasets accessible via MDAnalysisData, regardless of the simulation
package or analysis code that they use. Users are encouraged to cite the
authors of the datasets.
MDAnalysisData does not store files and trajectories. Instead, it
provides accessor code to seamlessly download (and cache) files from archives.
Outline
When you contribute data then you have to do two things
deposit data in an archive under an Open Data compatible license (CC0 or CC-BY preferred)
write accessor code in
MDAnalysisDataThe accessor code needs the stable archive URL(s) for your files and SHA256 checksums to check the integrity for any downloaded files. You will also add a description of your dataset.
Note
We currently have code to work with the figshare archive so choosing figshare will be easiest. But it should be straightforward to add code to work with other archive-grade repositories such as zenodo or DataDryad. Some universities also provide digital repositories that are suitable. Open an issue in the Issue Tracker for supporting other archives.
Step-by-step instructions
To add a new dataset deposit your data in a repository. Then open a pull request for the https://github.com/MDAnalysis/MDAnalysisData repository. Follow these steps:
STEP 1: Archival deposition
Deposit all required files in an archive-grade repository such as figshare.
Note
The site must provide stable download links and may not change the content during download because we store a SHA256 checksum to check file integrity.
Make sure to choose an Open Data compatible license such as CC0 or CC-BY.
Take note of the direct download URL for each of your files. It should be
possible to obtain the file directly from a stable URL with curl or
wget. As an example look at the dataset for
MDAnalysisData.adk_equilibrium at DOI 10.6084/m9.figshare.5108170 (as
shown in the figure below). Especially note the
download links of the DCD trajectory
(https://ndownloader.figshare.com/files/8672074) and PSF topology files
(https://ndownloader.figshare.com/files/8672230) as these links will be needed
in the accessor code in MDAnalysisData in the next step.
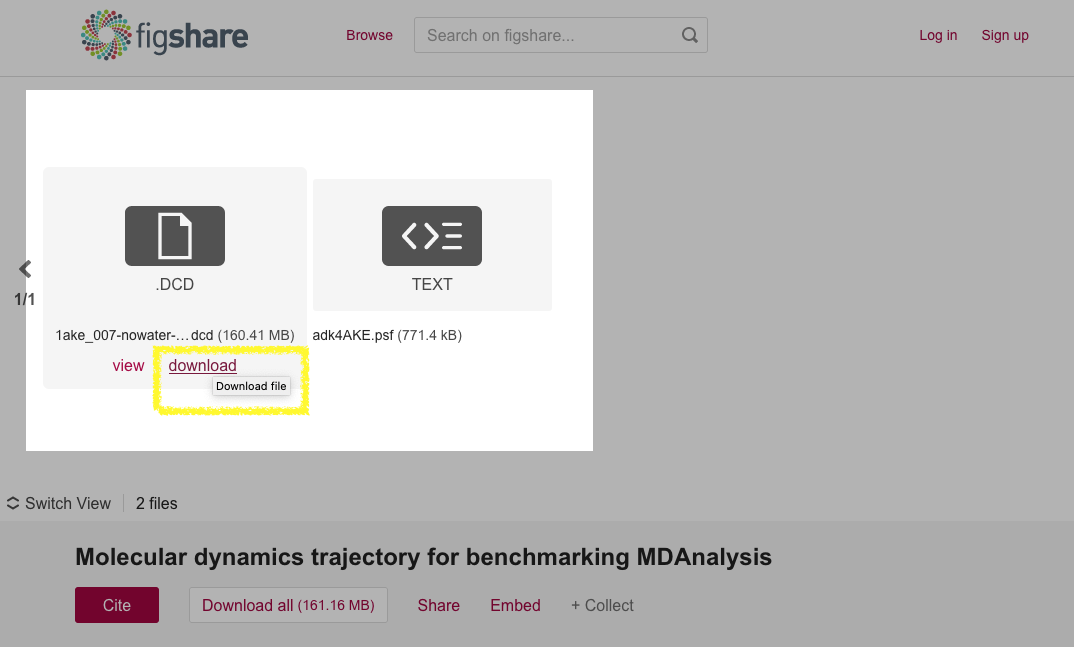
The AdK Equilbrium dataset on figshare DOI 10.6084/m9.figshare.5108170, highlighting the deposited trajectory and topology files. The download URLs are visible when hovering over a file’s image.
STEP 2: Add code and docs to MDAnalysisData
Add a Python module
{MODULE_NAME}.pywith the name of your dataset (where{MODULE_NAME}is just a placeholder). As an example see MDAnalysisData/adk_equilibrium.py, which becomesMDAnalysisData.adk_equilibrium). In many cases you can copy an existing module and adapt:text: describe your dataset
NAME: name of the data set; will be used as a file name so do not use spaces etcDESCRIPTION: filename of the description file (which contains restructured text format, so needs to have suffix.rst)ARCHIVE: dictionary containingRemoteFileMetadatainstances. Keys should describe the file type. Typicallytopology: topology file (PSF, TPR, …)
trajectory: trajectory coordinate file (DCD, XTC, …)
structure (optional): system with single frame of coordinates (typically PDB, GRO, CRD, …)
name of the
fetch_{NAME}()function (where{NAME}is a suitable name to access your dataset)docs of the
fetch_{NAME}()functioncalculate and store the reference SHA256 checksum as described below
Add a description file (example: MDAnalysisData/descr/adk_equilibrium.rst); copy an existing file and adapt. Make sure to add license information.
Import your
fetch_{NAME}()function in MDAnalysisData/datasets.py.from .{MODULE_NAME} import fetch_{NAME}
Add documentation
{NAME}.rstin restructured text format under docs/ (take existing files as examples) and append{NAME}to the secondtoctreesection of the docs/index.rst file... toctree:: :maxdepth: 1 :caption: Datasets :hidden: adk_equilibrium adk_transitions ... CG_fiber {NAME}
If your data set does not follow the same pattern as the example above (where
each file is downloaded separately) then you have to write your own
fetch_{NAME}() function. E.g., you might download a tar file and then
unpack the file yourself. Use scikit-learn’s sklearn/datasets as examples,
make sure that your function sets appropriate attributes in the returned
Bunch of records, and fully document what is
returned.
RemoteFileMetadata and SHA256 checksum
The RemoteFileMetadata is used by
_fetch_remote() and it will check file integrity by
computing a SHA256 checksum over each downloaded file with a stored reference
checksum. You must compute the reference checksum and store it in your
RemoteFileMetadata data structure for each file.
Typically you will have a local copy of the files during testing. You can
compute the SHA256 for a file FILENAME with the following code:
python import MDAnalysisData.base
print(MDAnalysisData.base._sha256(FILENAME))
or from the commandline
python -c 'import MDAnalysisData; print(MDAnalysisData.base._sha256("FILENAME"))'
where FILENAME is the file that is stored in the archive.